Controlled 'One-Cell-One-Bead' Encapsulation in Droplets
Brief Description
The invention relates to a technique using a microfluidic device to perform co-encapsulation of samples in droplets and sorting of said droplets. This technique can achieve a one-one-one encapsulation efficiency of about 80% and can significantly improve the droplet sequencing and related applications in single cell genomics and proteomics.
.Full Description
Droplet-based single cell assays are based on the ability to encapsulate and confine single cells in individual droplets and enable high-efficiency genome wide expression profiling.Encapsulation of a single cell and bead reagent is of critical importance when conducting droplet-based analysis of single cells. Currently, single cell encapsulation in droplets is randomly performed and not well controlled. One-cell-one-bead encapsulation efficiency is limited by an uncontrolled random droplet encapsulation of cells and beads (0.1% or 1 in 1000 droplets). Such a low droplet encapsulation efficiency hinders the realization of high throughput droplet-based experiments. Hence there is a need for improved microfluidic device for encapsulation in single droplets and sorting thereof.
UCI researchers have developed a microfluidic droplet encapsulation method to efficiently encapsulate one-cell and one bead in a single droplet. By designing a microfluidic device with controllable fluid flow and a high force liquid-liquid interface, one-cell one-bead droplet encapsulation efficacy can be well controlled. The invention features a two-step technique to achieve high efficiency cell indexing coupled with down-stream sorting (Figure 1). In the first step, >30% cell encapsulation can be achieved by an interfacial shearing method utilizing laminar flows and high shear liquid-liquid interface at a microfluidic junction. A sorting module is incorporated as a second step to improve the encapsulation efficiency further, by up to 80% by removing empty droplets and/or droplets that do not contain the desired number of cells, beads or both.
Utilizing the invented technology, it has been shown that a significant increase in droplet encapsulation efficiency can be readily achieved. While existing techniques only provide random encapsulation governed by Poisson statistics, the improved encapsulation efficiency in this invention enhances the ability to perform single cell genomics and proteomics on a microfluidic platform.
Figure 1
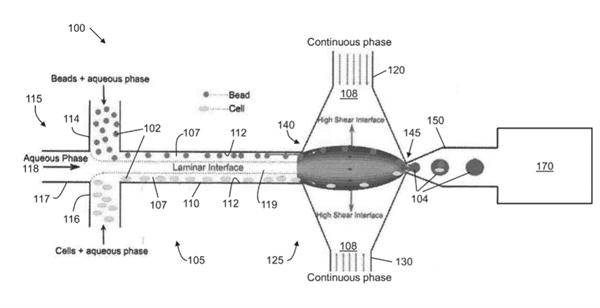
Figure 1 shows a schematic of high efficiency cell indexing in one-cell-one-bead droplets (1-1-1). Beads and cells introduced from upper and lower inlets self-assemble along the channel wall while moving toward high shear interfaces. At the droplet generation junction, both the beads and cells get pulled toward the high shear interface symmetrically from both the channel boundaries resulting in one-one-one encapsulation. A sorting module is incorporated downstream to sort out empty droplets from the bead-cell droplets.
State Of Development
Inventors have completed the experimental and validation phase with a working prototype. As the next step, the inventors plan to optimize device parameters in hopes to commercialize or license the technology.
Features/Benefits
Modular microfluidic technology that is capable of encapsulating one cell and one bead in one droplet
Increased droplet encapsulation efficiency
Improved droplet throughput for sequencing applications including single cell genomics and proteomics.
Suggested uses
- Improving pharmacological assay throughput by efficiently generating droplets that encapsulate a single cell and bead
- Capture specific proteins and antigens from single cells using functionalized beads
Patent Status
| Country | Type | Number | Dated | Case |
| United States Of America | Published Application | 20200108393 | 04/09/2020 | 2017-814 |
Additional Patent Pending
Contact
- Alvin Viray
- aviray@uci.edu
- tel: View Phone Number.
Inventors
- Lee, Abraham P.
Other Information
Additional Technologies by these Inventors
- New Microwell Plate Configurations to Increase Microwell Density
- Microfluidic device for multiplex diagnostics / Microfluidic devices and methods
- Microfluidic Device for Cell Separation Using Dielectrophoresis and/or Magnetohydrodynamics
- On-Demand Cell Encapsulation Using On-Demand Droplet Generation and Impedance-based Detection
- High throughput and precision cell sorting
- High-throughput Microfluidic Research Platform for Performing Versatile Single-Cell Molecular Timed-Release Assays within Droplets
