METHODS OF PRODUCING RIBOSOMAL RIBONUCLEIC ACID COMPLEXES FOR DIRECT RNA SEQUENCING
Background
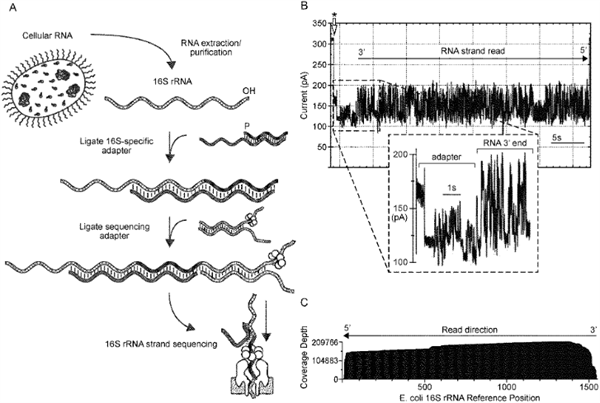
Long read nanopore sequencing can directly sequence RNA molecules, including rRNA, and result in full-length RNA sequences. rRNA sequencing is particularly useful for identifying microbes and full-length rRNA sequencing can identify microbes with post transcriptional modifications that confer antibiotic resistance. Such post transcriptional modifications are invisible to amplification based sequencing or other sequencing techniques that require reverse transcription.
Before this technology was developed, there were few if any efficient methods for preparing rRNA libraries for direct RNA sequencing, particularly for microbial identification in either a clinical or an environmental setting.
Technology Description
The methods involve hybridizing a probe complement oligonucleotide with an oligonucleotide probe. The oligonucleotide probe has one region that features a degenerate sequence complementary to the 3' region of rRNA and another region complementary to the probe complement oligonucleotide. A sequencing adapter is ligated to the probe complement oligonucleotide and the oligonucleotide probe. These form a complex that can then be directly sequenced in a long-read sequencer such as a nanopore sequencer. The patent includes claims to probes, libraries of complexes, other compositions, and kits.
Applications
- Direct rRNA sequencing in long read sequencing reactions.
- rRNA library preparations
- Identification of microbes in complex systems (clinic or environmental)
- e.g. 16S rRNA sequencing
Advantages
- Issued patent with broad actionable claims
- Efficient and simple system
Intellectual Property Information
| Country | Type | Number | Dated | Case |
| United States Of America | Issued Patent | 11,578,362 | 02/14/2023 | 2017-258 |
| United States Of America | Published Application | 20230295714 | 09/21/2023 | 2017-258 |
Related Materials
Contact
- Jeff M. Jackson
- jjackso6@ucsc.edu
- tel: View Phone Number.
Other Information
Keywords
ribosome, nanopore, phylogeny, taxonomy, oligonucleotide, RNA, sequencing
