Barcoded Solid Supports and Methods of Making and Using Same
Background
Barcodes are identifiable nucleic acid sequences that can be coupled to a target nucleic acid, either directly or indirectly. Doing so assists in analyzing the nucleic acids of interest. There are currently methods for introducing barcodes to long DNA molecules. However, long DNA molecules can be difficult to isolate. For example, long DNA molecules cannot be recovered from formalin-fixed-paraffin-embedded (FFPE) samples, but such samples are the major source of patient tumor DNA. There is a need for more efficient methods of using barcodes in haplotype phasing and other applications.
Barcoded nucleic acids on solid supports are used in a number of applications such as genome scaffolding, haplotype phasing, and single cell transcriptomics. Current methods involve the use of DNA amplification or chemical synthesis. These techniques are error prone and cost prohibitive.
Technology Description
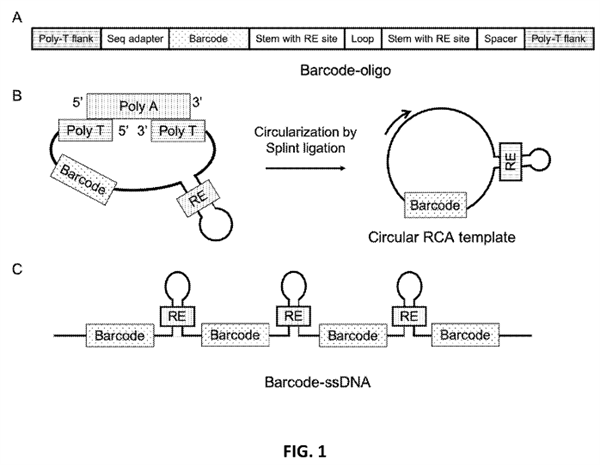
Researchers at UC Santa Cruz have developed ways to make barcoded solid supports. A linear nucleic acid with a barcode and a stem-loop forming region is circularized. This circular template is then used to make a concatemer via rolling circle amplification. The concatemer has tandem repeats (or linked units) of the barcode and stem-loop structure. Each unit of the concatemer can include additional domains, regions, or structures. For example, a sequencing adapter that is compatible with a sequencing platform such as Illumina® can be included. The concatemer is then disposed on a solid support, such as a bead. Both the concatemer and the solid support have binding members with affinity for each other. For example, the concatemer has binding members such as biotin, and the solid support has binding members as such as streptavidin so that the concatemer can be disposed on the solid support.
The barcoded solid support has several stem-loop structures extending from the surface of the solid support. The stem-loop structures can be cleaved to form stem structures with ends, such as overhangs or blunt ends, that are compatible with target nucleic acids. Target nucleic acids can be attached directly to the stem structures or attached via unique molecular identifiers.
These barcoded solid supports can be used in many downstream applications.
Applications
- haplotype phasing
- nucleic acid sequencing
- diagnostics
- structural variant identification
- topologically associated domain identification
- chromatin architecture
- genome assembly
- single cell genome sequencing
- HLA typing
- exome capture
- mitochondrial DNA enrichment
- custom gene panels
- transcriptome analysis
- single cell RNA sequencing
- targeted RNA sequencing
- metatranscriptomics
- biome analysis
- forensics
- linked-read sequencing
Advantages
- cost efficient
- adaptable to existing sequencing systems
- simple design
Intellectual Property Information
| Country | Type | Number | Dated | Case |
| United States Of America | Published Application | 2020038580 | 12/10/2020 | 2018-681 |
Contact
- Jeff M. Jackson
- jjackso6@ucsc.edu
- tel: View Phone Number.
Inventors
- Green, Richard E.
- Sundararaman, Balaji
Other Information
Keywords
concatemer, rolling circle amplification, haplotype phasing, barcode, bead
