Molecular And Computational Biology Methods For Improving Nanopore Sequencing Technology
Background
Long read sequencing (e.g. nanopore sequencing) involves a tradeoff between the length of the DNA fragment sequenced, which allows for greater ease of data assembly relative to massively parallel sequencing technologies (e.g. Illumina (R) sequencing) and accuracy of individual base calls.
This technology takes advantage of the long read capabilities of nanopore sequencing to improve the accuracy of reads of highly variable nucleic acid species, including cDNAs, and which can be highly variable due to alternative RNA splicing.
Technology Description
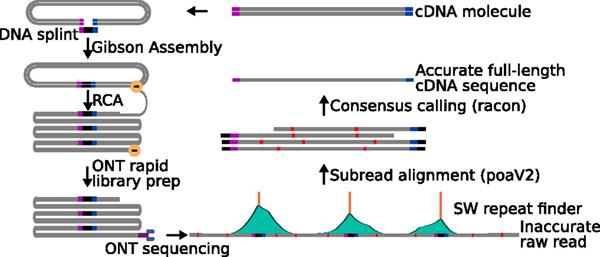
This technology incorporates both wet lab (concatamer formation by rolling circle amplification) and software (sequence analysis).
The technology involves producing a circularized DNA library via a Gibson Assembly that includes a full-length cDNA and a known heterologous sequence. The Gibson Assembly assures that only full-length cDNA's are included in the library. A rolling circle amplification is performed using the circularized DNA as template. This produces a concatemer or repeating segments of the full-length cDNA and the known heterologous sequence.
A raw sequencing read of the concatemer is obtained using long read sequencing (e.g. nanopore sequencing). The repeating segments in the raw sequencing read can be identified via a Smith-Waterman self-self alignment. From that, a consensus sequence of the full-length cDNA based on the sequences of the repeating segments can be produced.
Applications
- Long read nucleic acid sequencing
- Single cell cDNA sequencing
Advantages
- Allows accurate identification of closely related species (e.g. splice variants)
- Allows identification of rare mutants
- Whole cDNA's can be assessed
- Can be used to quantify RNA expression
Intellectual Property Information
| Country | Type | Number | Dated | Case |
| United States Of America | Issued Patent | 12,378,603 | 08/05/2025 | 2018-406 |
Additional Patent Pending
Related Materials
Contact
- Jeff M. Jackson
- jjackso6@ucsc.edu
- tel: View Phone Number.
Other Information
Keywords
Long Read Sequencing - improved accuracy, Gibson Assembly, Smith-Waterman self-self alignment, Full-length cDNA sequencing, Single Cell Sequencing, Rolling Circle Amplification
