Reading The 5 Prime End Of Eukaryotic Poly(A) Rna Molecules
Background
Nanopore sequencing requires a processive motor or other element to control the rate of RNA movement in single nucleotide steps through the nanopore sensor. The control element is typically situated several nucleotides from the sensor, therefore it necessarily releases before the end of the native RNA strand reaches the sensor. Thus, the bases along that terminal interval cannot be sequenced using conventional nanopore strategies.
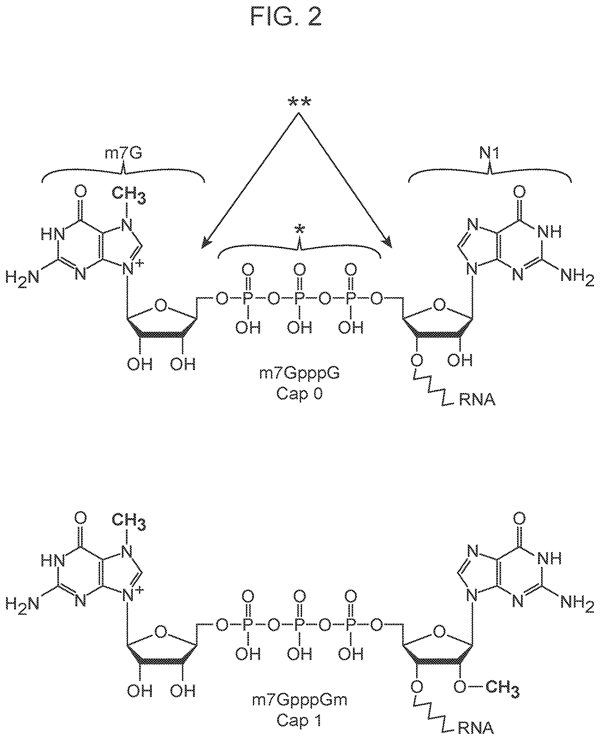
Furthermore the nucleotide sequence near that end in many eukaryotic RNAs is not typical. An important example is polyadenylated (poly A) RNA which often bears a 7 methylguanosine cap at the 5 prime end. The linkage between this modied cap and its neighbor is inverted, i.e. the two nucleotides are connected via the 5 prime carbons of their ribose sugars through triphosphate linker, rather than by a typical 5 prime to 3 prime linkage via a phosphodiester bond.
Technology Description
An extended adapter attached at the 5 prime end of polyA RNA bearing 7 methylguanosine allows for ionic current patterns to be measured through and past this 5 prime end. Features unique to this strand end including the triphosphate linker, the 5 prime to 5 prime linkage, and non biological components of the linker give unique ionic currents that could not predicted from prior art. These unanticipated ionic current features are important for identifying the 5 prime end of the RNA and for sequencing that 5 prime end at single nucleotide precision.
Applications
- Identifying the biological 5′ end of poly(A) RNA molecules
- Determination of transcription start sites
- Determination of full length RNA isoforms,
- Identification of nanopore poly(A) RNA reads that are not full length
- Identifying pre-processed RNA transcripts
- Detecting modifications in the nucleotides proximal to the 5′ cap
- Optimizing RNA current trace information for a specific read using the existing 3 prime poly(A) tail, features built into the 3 prime adapter, and features built and adapter attached to the prime end at the m7G cap
- Mapping the distance between the sensor in the pore and the RNA binding site in the motor enzyme
Advantages
Prior to this work, the 5 prime end of eukaryotic polyA RNA could not be detected using nanopore sequencing. An extended adapter attached at the 5 prime end of polyA RNA bearing 7 methylguanosine allows for ionic current patterns to be measured through and past this 5 prime end.
Intellectual Property Information
| Country | Type | Number | Dated | Case |
| United States Of America | Issued Patent | 12,195,794 | 01/14/2025 | 2018-688 |
| United States Of America | Published Application | 20250101507 | 03/27/2025 | 2018-688 |
Related Materials
Contact
- Jeff M. Jackson
- jjackso6@ucsc.edu
- tel: View Phone Number.
Inventors
- Akeson, Mark A.
- Jain, Miten
- Mulroney, Logan
- Olsen, Hugh E.
Other Information
Keywords
RNA sequencing, Long read sequencing, Nanopore sequencing, RNA-Seq, Full length mRNA sequencing, poly-A RNA, 5' end sequencing
