Simplified Workflow For Hybridoma Antibody Sequencing
Background
Before recombinant antibody expression plasmids can be designed, sequncing of the antibody light and heavy chain variable regions is necessary. Several other methods of sequencing antibody variable regions are available. Some involve high throughput RNA sequencing. These techniques are unavailable to many labs; they require the preparation of RNA-seq libraries, and computational analysis. As a result, the cost of performing such techniques is substantial and with sequencing cores being oversubscribed, turnaround can be as long as weeks to months.
Other methods involve PCR and Sanger sequencing. However PCR amplification of variable regions results from difficulties in generating universal primers that can amplify any given variable region - particularly given the inherent low sequence identity in the 5' leader sequence of antibody light chains and heavy chains upstream of the variable regions. Sometimes degenerate primers can be used, but amplification success rate is only 80-90% due to non-specific priming and/or failure to prime at all. In addition, there is a significant risk that the variable regions of the parental myeloma line can amplify using the degenerate primers.
5' RACE (rapid amplification of 5' cDNA ends) can also be used, but mRNA degradation, cDNA purification and poly-A addition between reverse transcription and PCR, makes the technique long and difficult to perform. Non degenerate primers can be used, but each variable region requires multiple amplification attempts with different primer sets as well as sequence validation using mass spectrometry. And with both of these methods, primer derived mutations can be introduced.
Mass spectrometry can be used to determine antibody variable regions, but these can result in ambiguous sequences because of isobaric residues such as isoleucine and leucine. But this method is time consuming, requires huge amounts of purified monoclonal antibody, is expensive and is inaccessible to most researchers.
This technology involves a template switch reverse transcription of hybridoma RNA with at least three chain specific RT primers - one for the kappa chain, one for the lambda chain, and at least one for the heavy chain (for efficiency, this can be limited to IgG in a first pass). These are amplified in three separate PCR reactions and sequenced using Sanger sequencing.
Technology Description
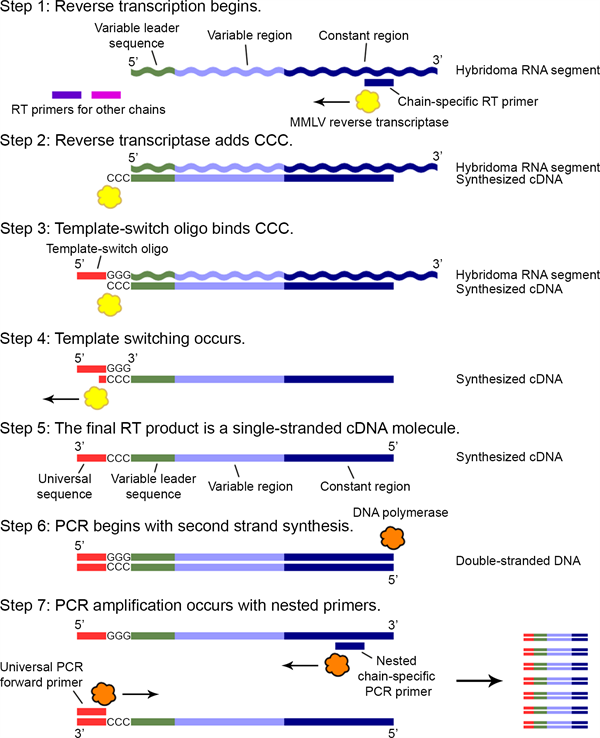
The process (outlined below) involves reverse transcription from a constant region specific primer (one for kappa, one for lambda, and at least one for the heavy chain). The reverse transcriptase adds a CCC sequence to the first cDNA strand. A template switch oligo with a GGG sequence hybridizes to the CCC sequence. Continued reverse transcription results in a single stranded cDNA complementary to the variable region plus part of the constant region. This single stranded cDNA can then be used as a PCR template. At least three different PCR reactions using a nested chain specific primer specific for kappa, lambda, and at least one heavy chain, result in PCR amplified variable regions. These can readily be sequenced using cheap and readily avialable Sanger sequencing.
Applications
- Sequencing V-regions in monoclonal antibodies from hybridomas
- Hybridoma quality control
- Hybridoma identification
Advantages
- Fast
- Simple
- Accurate
- Easily accessed by any lab (works with Sanger sequencing)
- User friendly - dozens of unsolicited positive comments by end users.
Intellectual Property Information
| Country | Type | Number | Dated | Case |
| United States Of America | Published Application | 2020039257 | 12/17/2020 | 2018-683 |
Related Materials
Contact
- Jeff M. Jackson
- jjackso6@ucsc.edu
- tel: View Phone Number.
Inventors
- DuBois, Rebecca M.
- Meyer, Lena
- Vollmers, Christopher
Other Information
Keywords
hybridoma sequencing, antibody v-region, heavy chain, light chain, kappa chain, lambda chain, IgG, template switch, sequencing, Sanger sequencing, RNA-seq, reverse transcription, hybridoma, monoclonal antibody, monoclonal antibody v-region sequencing
Categorized As
Additional Technologies by these Inventors
- Methods For Production Of The Porcine Astrovirus 4 Capsid Spike Antigen And Its Use In Serological Assays And Vaccines
- Glycoengineering Of The Foldon Protein Trimerization Domain To Shield It From Antibody Immune Responses
- Human Astrovirus Neutralizing Monoclonal Antibody Sequences
- TMI-seq: Tn5 Transposase Mediated Production of Complex Libraries for Short Read Sequencing
- Methods To Rapidly Measure Antibodies And Other Biomolecules In Clinical Specimens Utilizing Biolayer Interferometry
